Sudin Bhattacharya

Contact
sbhattac@msu.edu
517-884-6952
View publications on MSU Scholars
IQ DIVISION – Systems Biology
Departmental AFFILIATIONS
ABOUT
Dr. Sudin Bhattacharya (he/him/his) is an Associate Professor in the departments of Biomedical Engineering and Pharmacology & Toxicology. He leads a lab that conducts research at the interface of computation and biology, using quantitative tools to study the signaling and transcriptional networks that regulate cell fate and its perturbation by environmental pollutants.
Dr. Bhattacharya joined MSU as an assistant professor in November 2015. Originally from Kolkata, India, Dr. Bhattacharya completed his undergraduate degree at Jadavpur University in India, his master’s degree at the University of Kentucky, and his Ph.D. at the University of Michigan, all in mechanical engineering. He completed his postdoc in computational biology at The Hamner Institutes in Research Triangle Park, North Carolina.
THE BHATTACHARYA LAB
The Bhattacharya Lab uses a variety of computational tools to address fundamental problems across multiple scales in biology and toxicology.
Like all fields in biology, toxicology is being revolutionized by new technologies like single cell genomics and artificial intelligence. We are applying a variety of predictive and mathematical modeling tools to understand how tissues, cells and gene regulatory networks are perturbed under exposure to potentially toxic chemicals.
MAJOR AREAS OF RESEARCH
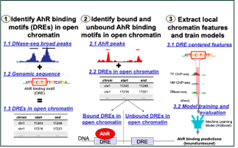
Prediction of DNA – transcription factor binding
Predicting genome-wide transcription factor binding sites on DNA is a challenging problem. We are using machine learning tools to address this problem for members of the bHLH-PAS family of transcription factors.
- Prediction of mammalian tissue-specific CLOCK-BMAL1 binding to E-box motifs, Marri et al 2023.
- Predictive Models of Genome-wide Aryl Hydrocarbon Receptor DNA Binding Reveal Tissue Specific Binding Determinants, Filipovic et al 2023.
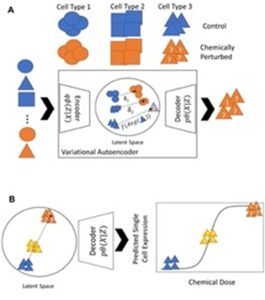
Modeling perturbations in single-cell gene expression
The space of human cell types and likely chemical treatments / doses is exponentially large. We are developing deep generative AI tools to extrapolate chemical dose-response across cell types, doses and chemicals.
- Generative Modeling of Single Cell Gene Expression for Dose-Dependent Chemical Perturbations, Kana et al, 2023.
Predictive modeling of cellular transitions in health and disease
We are integrating and analyzing bulk and single-cell gene expression data as part of a multi-lab project investigating the role of adipose tissue in cardiovascular disease.
- Phenotypic Changes in T Cell and Macrophage Subtypes in Perivascular Adipose Tissues Precede High-Fat Diet-Induced Hypertension, Kumar et al, 2021.
- Blood pressure changes PVAT function and transcriptome: use of the mid-thoracic aorta coarcted rat, Contreras et al, 2020.
Talk at MSU Physiology (2018):

