Michigan State University has landed a $2.35 million National Institutes for Health R01 grant to investigate the effect of environmental pollutants and toxins on activation of genes that have effects on the liver and immune system.
Human exposure to the highly toxic compound 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) and other dioxin-like chemicals remain a public health concern, especially for vulnerable populations like infants. While most biological effects of TCDD are known to be mediated by the aryl hydrocarbon receptor (AHR), the identity of specific genes connecting AHR activation to toxic outcomes of TCDD exposure remains unknown.
Sudin Bhattacharya, an assistant professor of Biomedical Engineering and an investigator at the Institute for Quantitative Health Science and Engineering, wants to identify these genes. By combining functional genomic experiments, computational modeling, and targeted epigenome editing, his research aims to map AHR-mediated gene regulatory networks and predict TCDD-induced changes in gene expression in mouse and human liver and B cells.
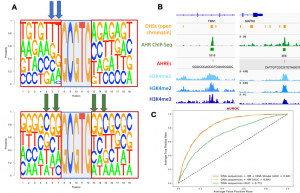
Classification of bound vs. unbound Aryl Hydrocarbon response elements (AHREs). (A) Sequence logos derived from 720 bound (top panel) and 800 unbound (bottom panel) AHREs. The shaded box in the middle of each panel denotes the invariant 5’-GCGTG-3’ core of the AHRE. (B) Overlap between DHS peaks (open chromatin) Aryl Hydrocarbon Receptor (AHR) ChIP-Seq peaks, bound AHREs, and histone modifications (HMs) H3K4me1, H3K4me2 and H3K4me3 in MCF-7 cells. (C) XGBoost machine learning model was able to classify bound from unbound AHREs. Inclusion of HMs improved area under the receiver operating characteristic curve (AUROC) from 0.71 to 0.84.
“Much remains unknown about how transcriptional regulatory proteins activated by industrial pollutants find their appropriate binding sites in the genome and regulate target genes,” explains Dr. Bhattacharya. “Using the AHR as an example of an inducible protein, we will identify mechanisms of genome-wide binding and target gene regulation both from neighboring and distal binding sites in liver and immune cells.”
The five-year R01 is a multi-PI grant in collaboration with Suresh Cuddapah from NYU Grossman School of Medicine. The team of scientists also includes Co-PIs Norb Kaminski, Tim Zacharewski, and Jianrong Wang from MSU.
Contacts: Sudin Bhattacharya, Saneeya Mir


